1.1 - ABOUT SEQEDITOR
SeqEditor is a Sequence Browser for the management and analysis of nucleotide and protein sequences of up to 300 megabases. This enables it to manage the largest chromosomes currently known. SeqEditor is an updated version of TIME, a former sequence editor (
Muñoz et al. 2011) also implemented as a sequence editor in the former release of GPRO (
Futami et al. 2011). The new version provides improved updates of the functions (translation of nucleotide sequences into proteins, editing of sequences, changes in the geometry and orientation of the sequences etc.) and also provides new utilities for the searching and filtering of sequences, ORFs and motifs, managing multiple files simultaneously, primer design, metrics inference, and more.
CITING SEQEDITOR:
Hafez A, Futami R, Arastehfar A, Daneshnia F, Miguel A, Roig FJ, Soriano B, Perez-Sánchez J, Boekhout T, Gabaldón T, Llorens C. 2020. SeqEditor: an application for primer design and sequence analysis with or without GTF/GFF files. Bioinformatics, btaa903,
https://doi.org/10.1093/bioinformatics/btaa903
CITING THE GPRO SUITE:
Futami R, Muñoz-Pomer A, Viu JM, Dominguez-Escribá L, Covelli L, Bernet GP, Sempere JM, Moya A, Llorens C. 2011. GPRO: the professional tool for management, functional analysis and annotation of omic sequences and databases. Biotechvana Bioinformatics: 2011-SOFT3,
http://bioinformatics.biotechvana.com/index.php/article/35
1.2 - VERSION and DOWNLOADS
SeqEditor is distributed as an installer for Windows 7 or later (64 bit), a self-extracting disk image for Mac OS X 10.6 or later (64 bit), and a compressed tarball file for Linux 2.6 kernel series or later (64 bit). You can download the latest version of these executables at this [
link]
1.3 - INSTALLING SEQEDITOR IN YOUR PC
Every software application of the GPRO suite is a standalone Java app that requires a minimum 2GB of RAM or higher as well as the installation of a Java JRE (Java Runtime Environment) version 6 or superior.
To check if you already have a JRE installed on your computer, open a command line interface and type:
Assuming that you have the java version 1.8.0_xx you should see a message like the following:
$ java -version
java version "1.8.0_92"
Java(TM) SE Runtime Environment (build 1.8.0_92-b14)
Java HotSpot(TM) 64-Bit Server VM (build 25.92-b14, mixed mode)
|
If you get a “Command not found” error message, it means that JRE is not installed. For installing JRE, go to the official JRE repository
here and download the version matching your operating system. Once installed, check again the output of java -version command on a command line interface. Sometimes, although the JRE is installed, it is not set at the path.
To install the Windows version
Download the
SeqEditor-win32.win32.x86_64.zip file and unzip it.
Then browse to the executable file "VariantSeq.exe" and execute/run it.
To install the Mac version
Download the
SeqEditor-macosx.cocoa.x86_64.zip file and unzip it
Then browse to the executable binary file "VariantSeqr.app" and execute/run it.
To install the Linux version
Download the
SeqEditor-linux.gtk.x86_64.zip file and unzip it
Then browse to the executable binary file "VariantSeq" and execute/run it.
1.4 - RAM ASSIGNATION TO YOUR PC
The amount of RAM memory assigned for SeqEditor can be tweaked in a configuration file named “SeqEditor.ini” that can be located inside SeqEditor application folder on
Linux and Windows computers.
On macOS computers, this configuration file can be found by right-clicking on SeqEditor.app → Show package contents → Contents → MacOS → SeqEditor.ini
Open the file with any plain text editor and locate these two following parameters:
- Xms1024m (Minimum allocated memory)
- Xmx2048m (Maximum allocated memory)
|
Change the values according to the amount of physical memory available on your computer. E.g. if your computer has 8GB of physical memory, it is recommended assigning Xms2048m and Xmx4096m for a better performance. If required, Xmx can be increased up to Xmx6144m for handling large datasets. Avoid using values near to the maximum available memory as it can lead to computer’s operating system unstability
1.5 - GETTING FAMILIAR
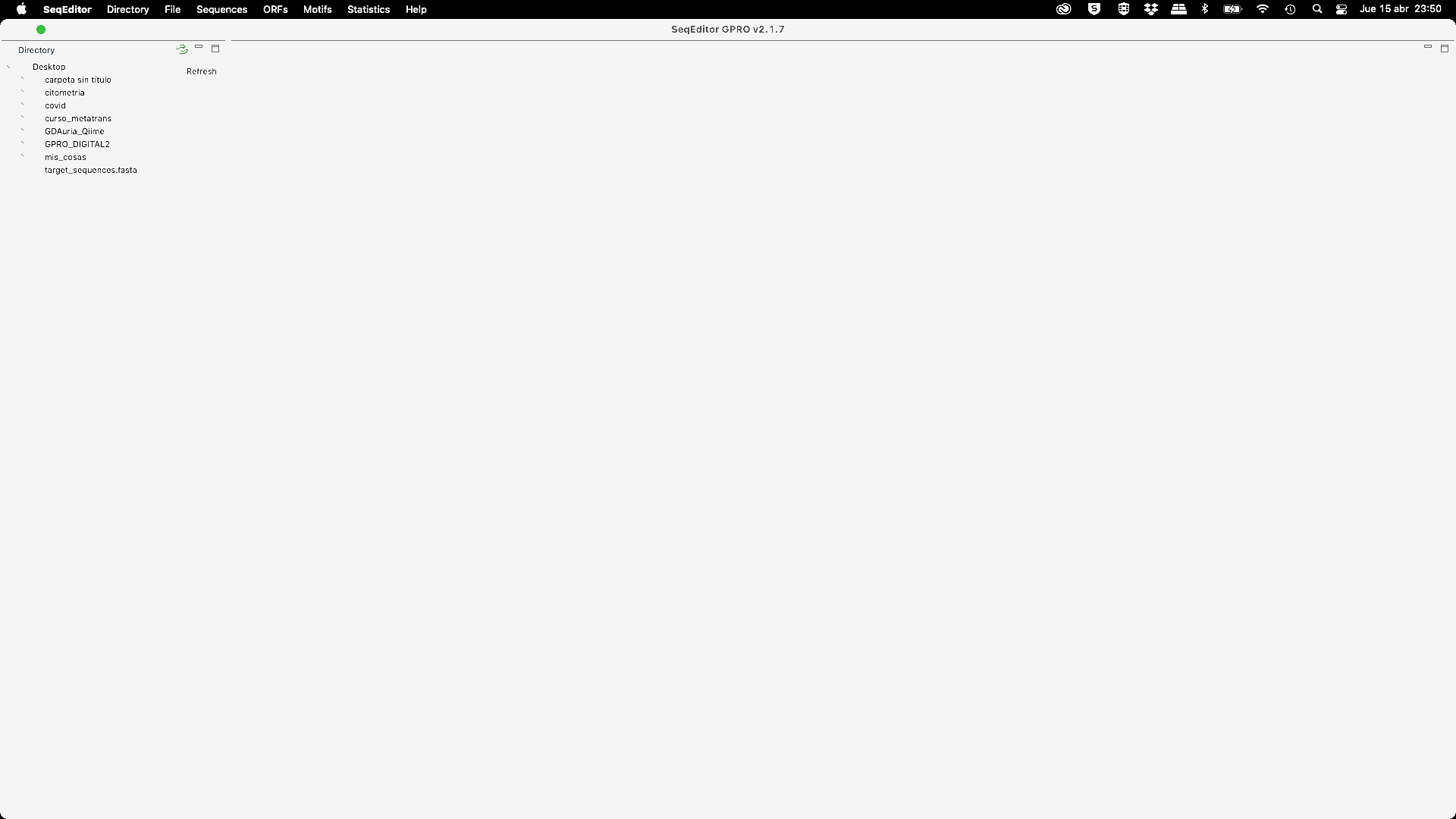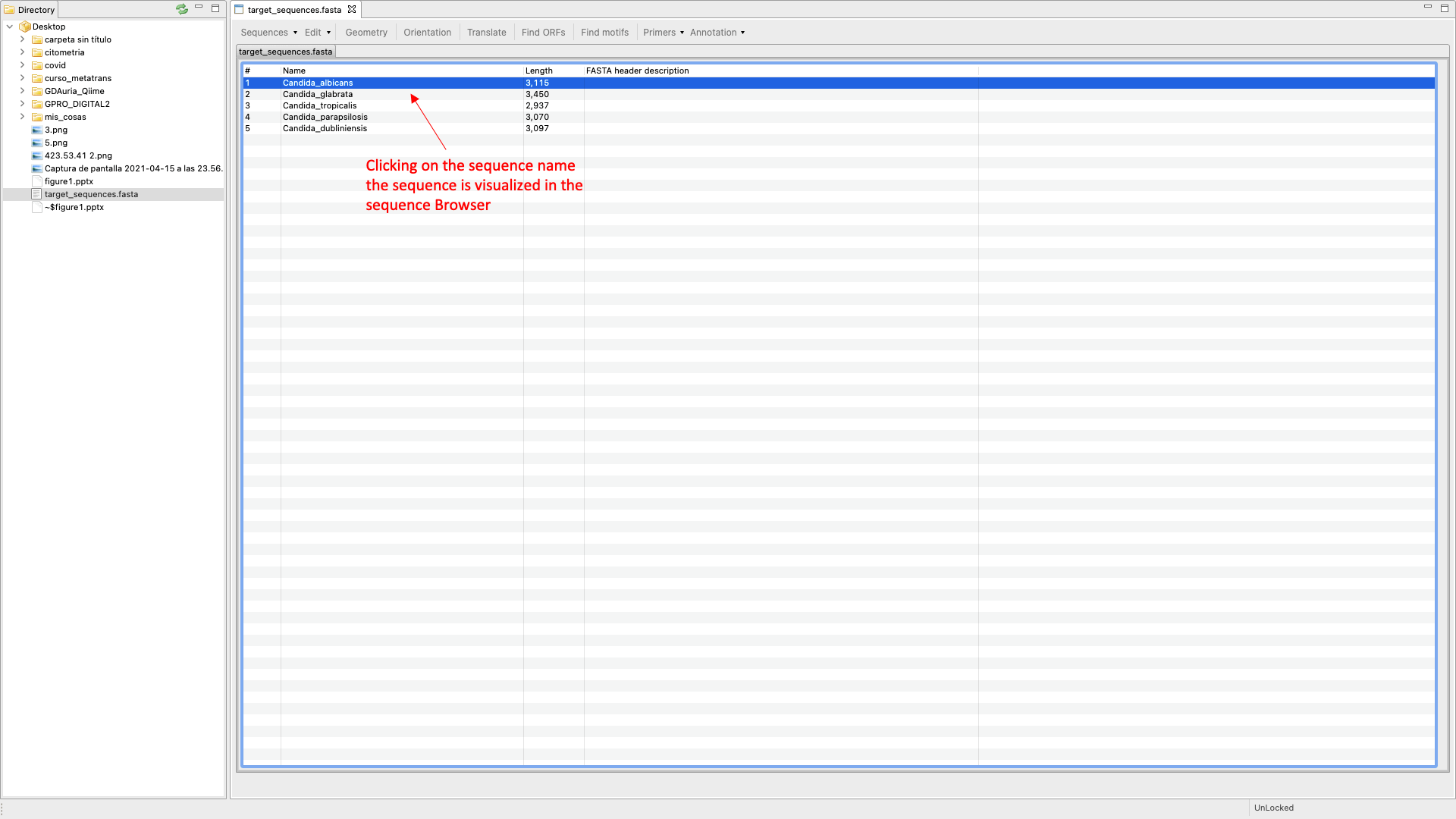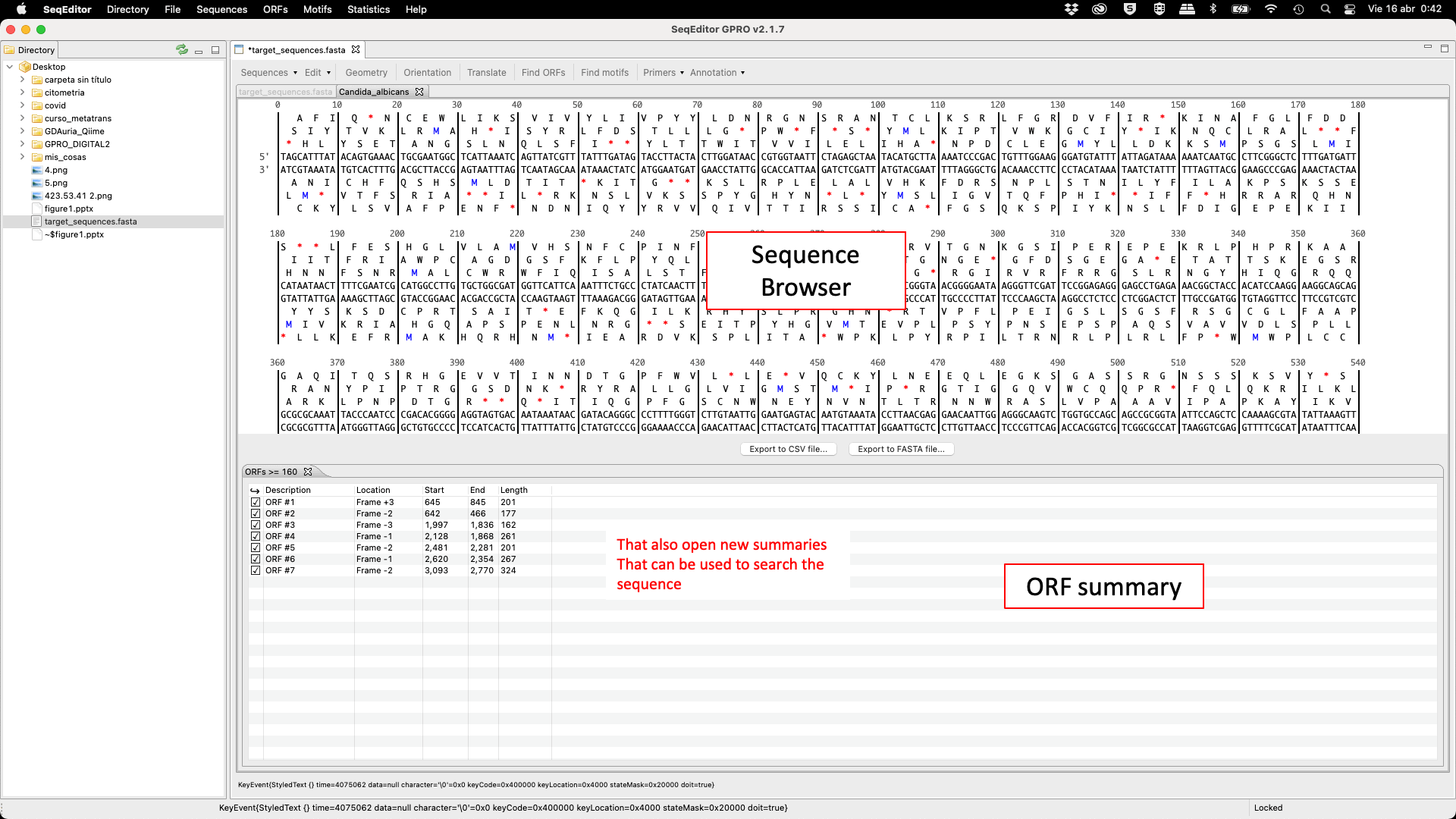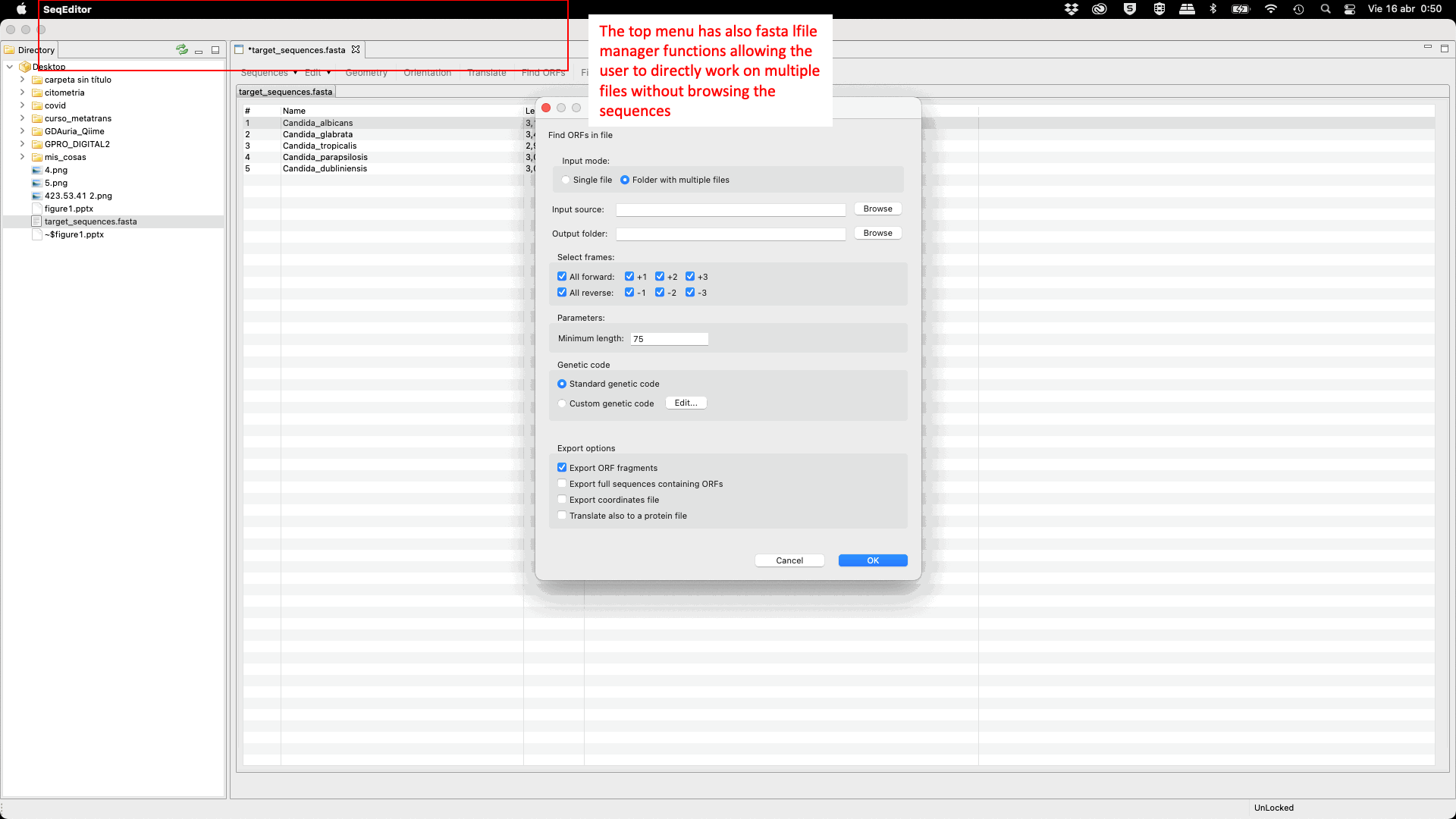
SeqEditor can work as a fasta manager or as a graphical sequence browser. Figure 1 shows a screenshot of SeqEditor, whose layout is composed of four main sections: “directory browser” (controlling the information that displayed in a directory listing), “top menu” (allowing the user to access the sequence browser or to work with several sequences or fasta files at the same time), “sequence browser” (to graphically manage the sequence analysis), and “browser menu” allowing the users to analyze the current opened sequence at the sequence browser. Additional interfaces are also available to run the analyses or display summaries for each browser’s task. These include a “fasta manager dialog” that can be opened with any of the Top menu options and allows the user to analyze multiple sequences in a file or several fasta files, the “fasta file summary viewer” that shows the sequences included in a fasta file and their lengths, the “ORF Finder summary viewer”, that shows the found ORFs in a specific sequence and their characteristics, and the ”Browser task dialog”, that can be accessed through the browser menu options and varies in accordance to those functions.
1.6 - MOUSE ACTIONS
SeqEditor allows different functions by clicking on the directory browser or the Sequence Browser. By right clicking anywhere in the directory space, a dialog will appear enabling all the standard actions that allow the creation and management of files and folders. From this space, files can be cut, copied, pasted, deleted, and renamed. By right clicking on the sequence browser you can unlock sequences for further editing or copy and paste a sequence.