DeNovoSeq is a Client-Desktop Application of the GPRO suite with custom dedication to manage and run pipelines and workflows for de novo reconstruction and annotation of new genome and transcriptome sequences using NGS data and a protocol based on the State-of-the-Art (Fig.1). The application is coupled with an infrastructure of server-side dependencies (pipelines, databases, and tools) that we distribute in a container that can be installed on a remote server or on a PC with sufficient RAM. The application also includes a File Transfer Protocol system (FTP) to facilitate the upload and download of files from the user’s computer to or from the server; a progress tracker (job tracking system), and two different execution modes (a “step-by-step” mode, and a “pipeline-like” mode).
The Step-by-Step mode is a procedure similar to those implemented in Galaxy (Afgan et al 2018) and others GUI-based solutions for NGS data analysis. This mode organizes the different steps of the protocol for de novo analysis (i.e. quality analysis, preprocessing, de novo assembly, gene prediction, annotation and functional analysis) into an intuitive menu providing a selection of command line interface (CLI) third party software for each step. At the same time each CLI tool has an interface implementation with distinct fields to declare the inputs and outputs files or for tuning the options and parameters provided by that tool. In contrast, the pipeline mode is a pipeline configuration system allowing the user to execute all the steps of a given protocol automatically one after the other. To this end the user just need to select a specific pipeline from a list, declare the experiment design, as well as, the input and output data, then configure the option and parameters and finally run the pipeline where the distinct analyses will be executed sequentially one after another.
In contrast, the pipeline mode is a pipeline configuration system allowing the user to execute all the steps of a given protocol automatically one after the other. To this end the user just need to select a specific pipeline from a list, declare the experiment design as well as the input and output data, then configure the option and parameters and finally run the pipeline where the distinct analyses will be executed sequentially one after another.
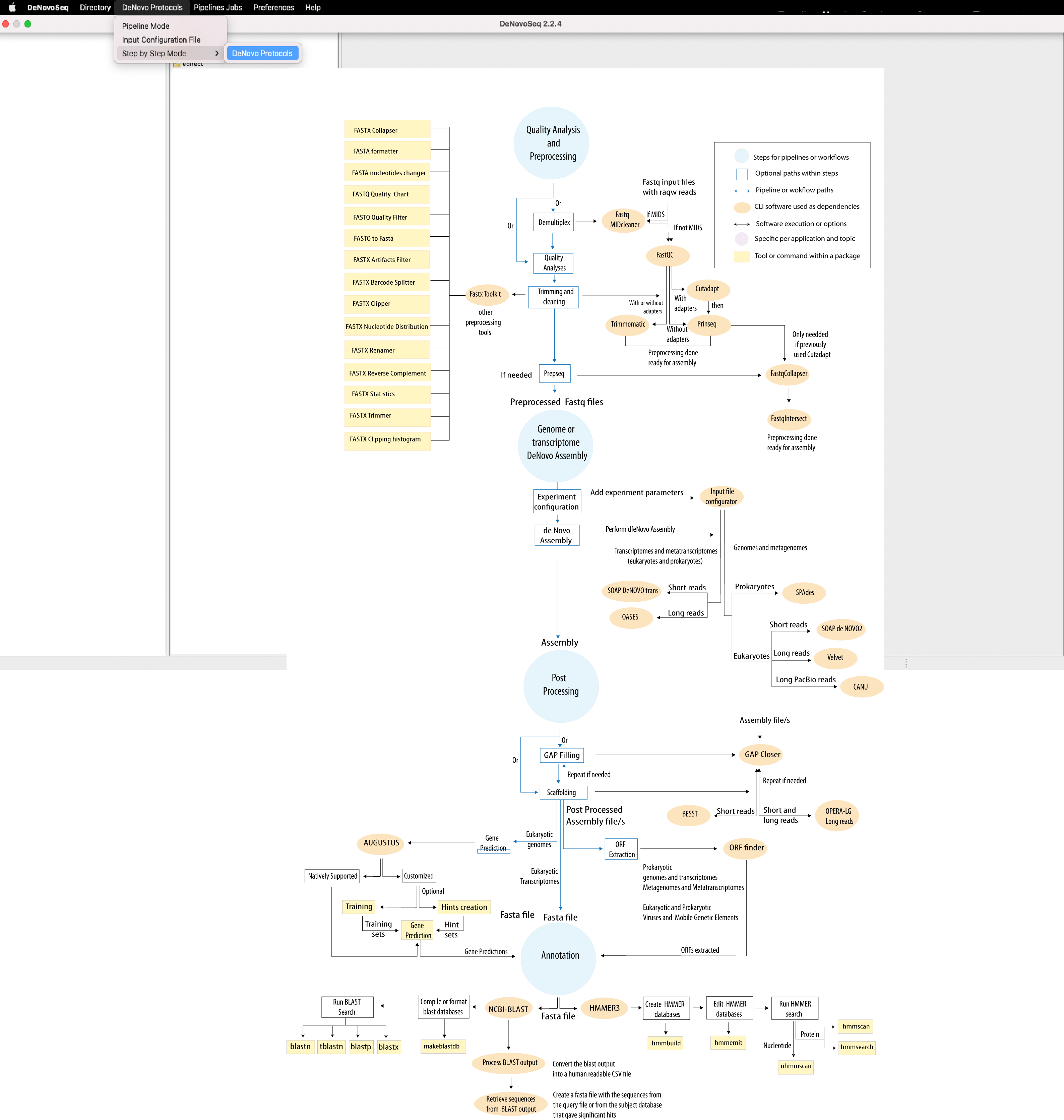
Figure 1: Bioinformatic protocol implemented in DeNovoSeq for reconstruction and annotation of new genome and transcriptome sequences for a first time and without the aid of a reference sequence template. The tool provides two execution modes (Step-By-Step and Pipeline-like). An interactive version of this protocol is available at GENIE our virtual assitant.
java -version |
$ java -version
|
DeNovoSeq-win32.win32.x86_64.zip file and unzip it.
Then browse to the executable file “DeNovoSeq.exe” and execute/run it.
DeNovoSeq-macosx.cocoa.x86_64.zip file and unzip it. Then browse to the executable binary file "DeNovoSeq.app" and execute/run it.
DeNovoSeq-linux.gtk.x86_64.zip file and unzip it. Then browse to the executable binary file “DeNovoSeq” and execute/run it.
DeNovoSeq is a Client Side + Server Side solution thus meaning that the application is coupled via API with a bioinformatic infrastructure called GPRO Server Side that contains all the dependencies needed by DeNovoSeq to execute the workflows and pipelines. These dependencies are scripts, databases and the following third party CLI software:
The GPRO Server Side can be installed in the PC of the user or in remote servers as a Cloud Computing resource. However, its installation is a complex task due to the amount of dependencies and requirements (besides of the CLI software) for installing and running this infrastructure. For this reason, we distribute the GPRO Server Side in a Docker container that can be easily installed for the user in a couple of steps. Indications for installation of the GPRO Server Side Docker are available here here.
As also shown in Figure 2 you can also check the option “Run GPRO server locally using Docker” to let you to automatically start the GPRO container each time you run DeNovoSeq (Also note that if you have this option checked you do not need to type the port). You can test if the app is connected to the Server Side clicking on the tab “Test connection settings”. Alternatively, if you install the Server Side manually (without the Docker) just add the IP of the remote server where the Server Side is hosted, add the port information (by default 22) and keep the Option “Run GPRO server locally using Docker” unchecked.
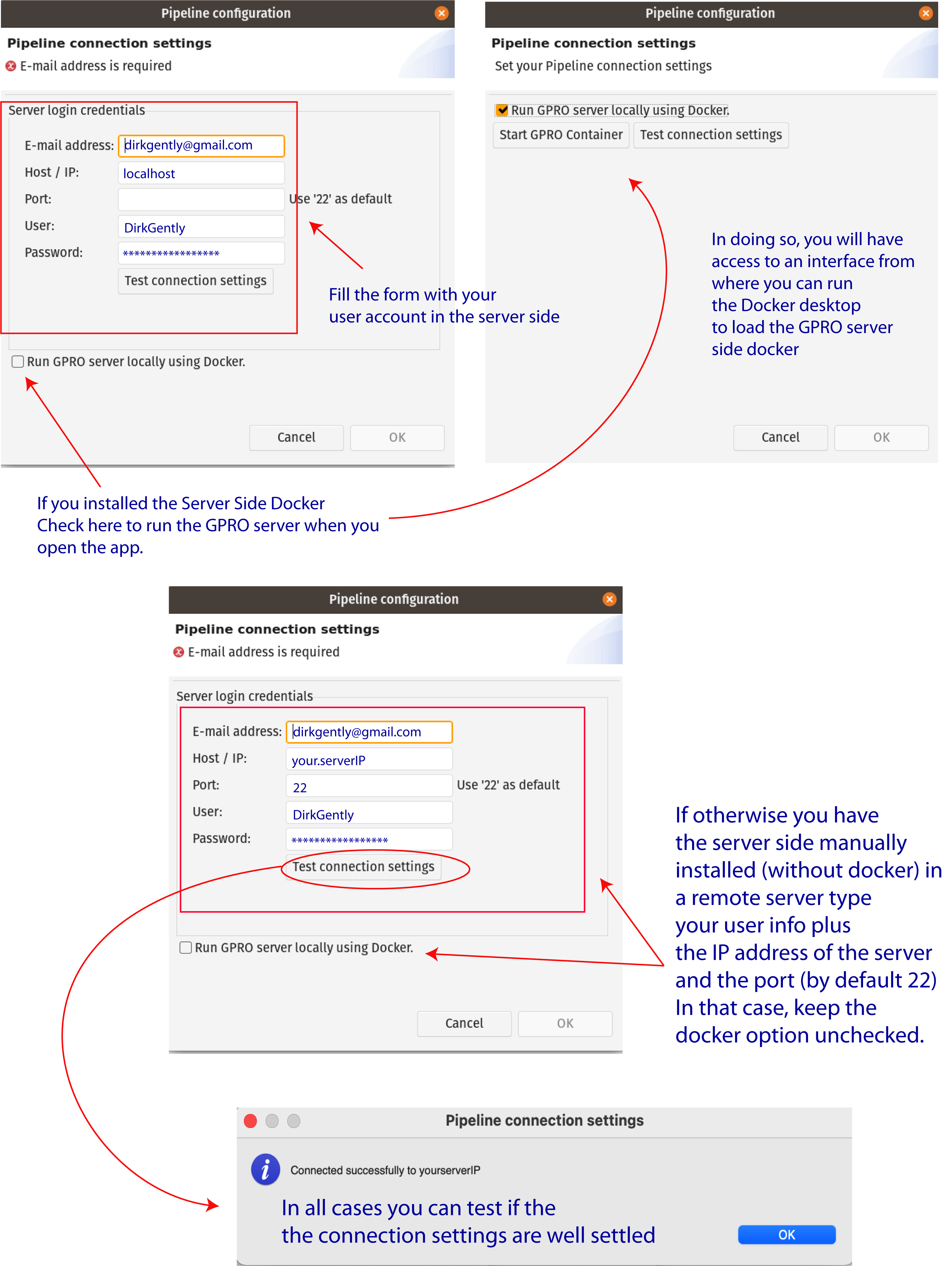
Figure 2: Server connection dialog.
[DeNovoSeq.app → Show package contents → Contents → MacOS → DeNovoSeq.ini].
Xms1024m (Minimum allocated memory)
|
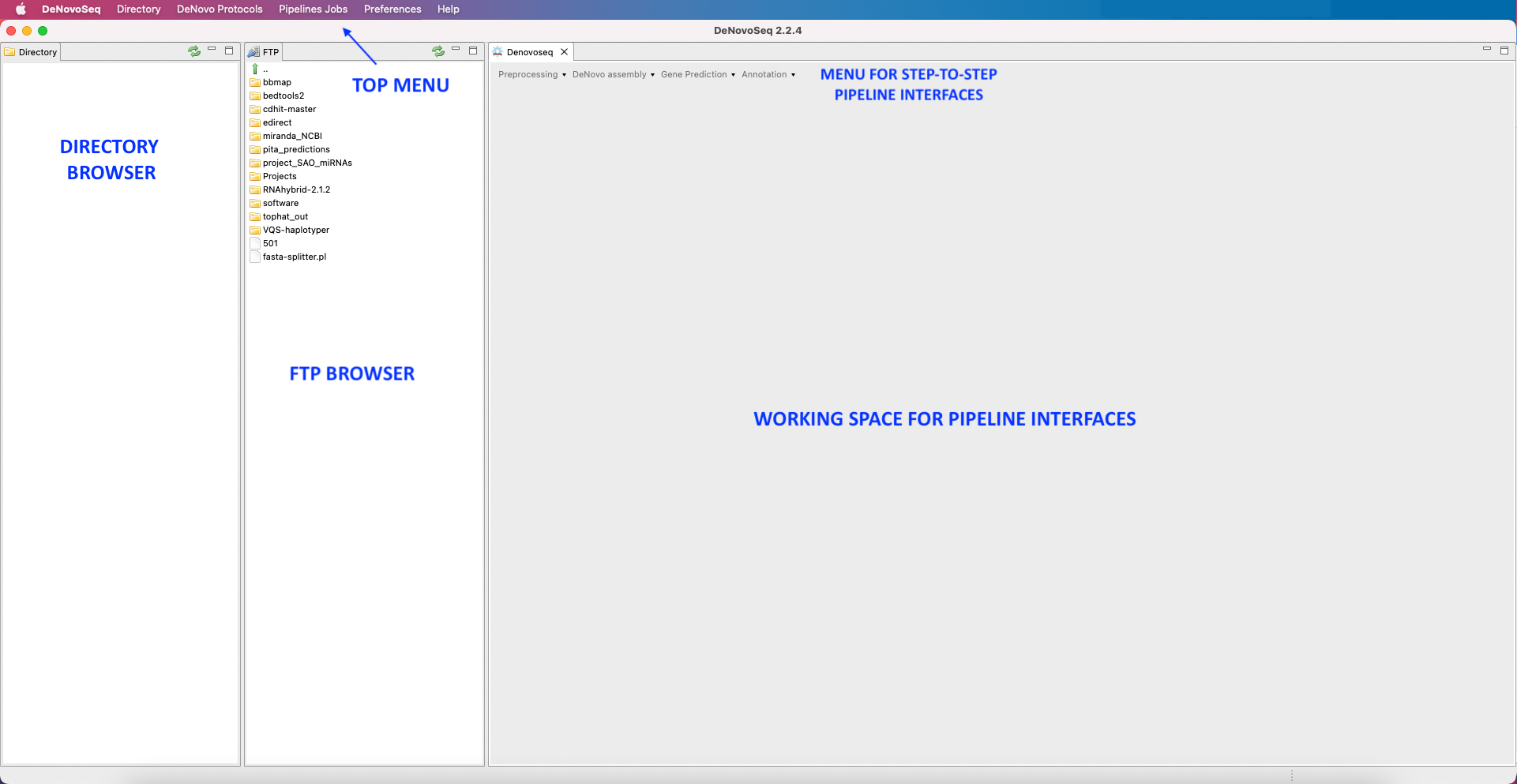
Figure 3: Main layout of DeNovoSeq. Both the Directory and FTP Browser windows can be either resized or masked by clicking on the window icons at the top right corner of their respective windows. All files and folders contained in either of these browsers can be managed manually using the mouse. Please keep in mind that the window views shown in this manual will change depending on the operative system used.
[Directory → Select directory folder ] : Selects a workspace from the directory browser.[Directory → Show ] : Shows the Directory Browser.[Directory → Hide ] : Hides the Directory Browser.[Denovo protocols -> Pipeline Mode ]: Configuration view to set up your assembly project and launch a the de novo assembly pipeline.[Denovo protocols -> Input Configuration file ] : Configuration the file used to declare number and type of samples, as well as type of experiment.[Denovo protocols -> Step-by-Step Mode → De Novo Protocols] : View of the available Protocols for de novo analysis in manual mode[Pipeline Jobs → Jobs Tracking System ] : For jobs tracking.[Pipeline Jobs → FTP Transfers ] : screen for tracking the jobs of the sFTP protocol. [Preferences → Pipeline Connections Settings] : Gives access to the server login details setup your user credentials for accessing the server.[Help → About DeNovoSeq ] : Technical details and copyright of DeNovoSeq, as well as links to the license and user manual.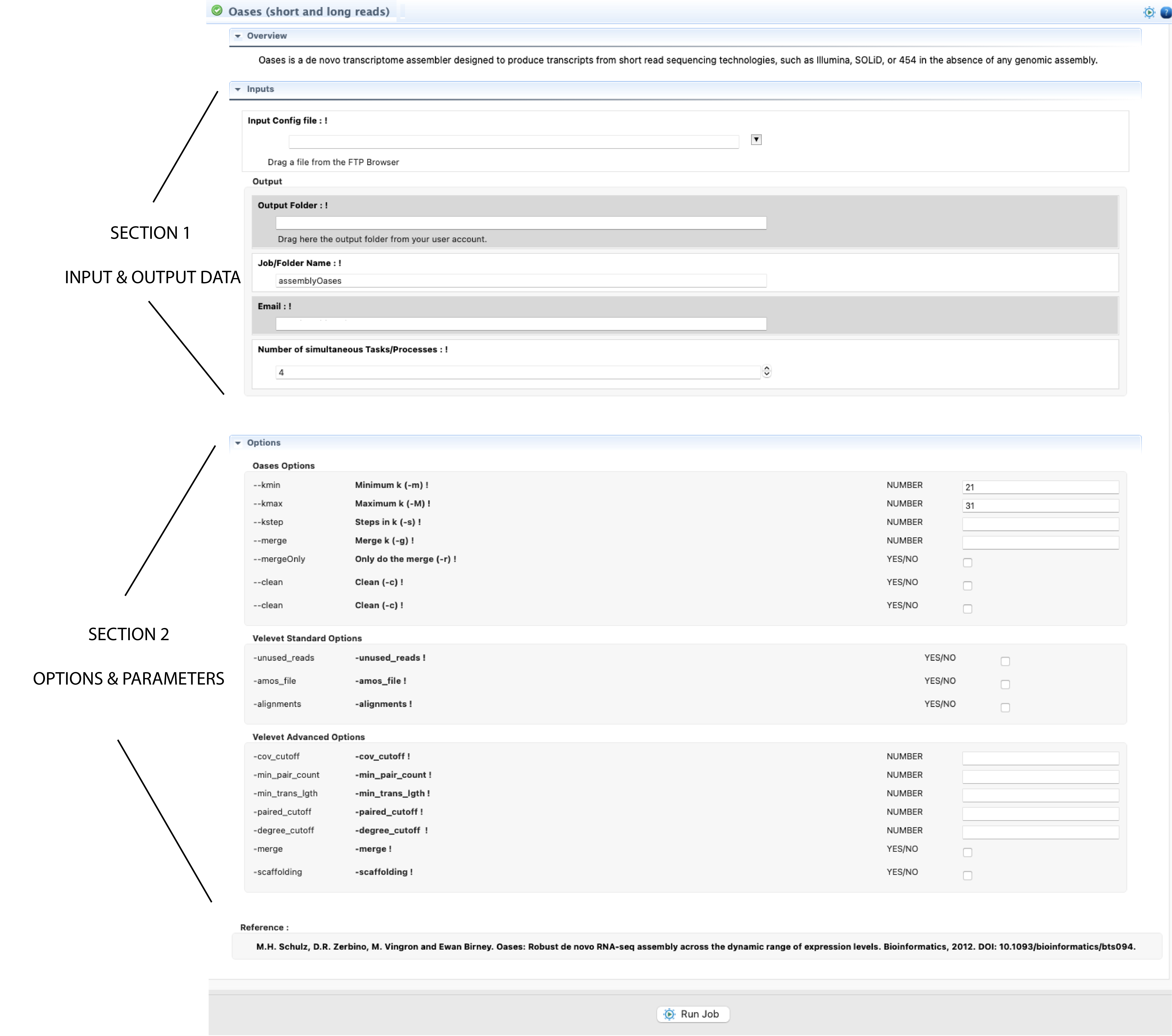
Figure 4: Example of interface for a CLI tool provided by the ”DeNovoSeq” Step-By-Step mode. The figure shows the interface for Oases.
From the FTP browser select the input file/s with the mouse and drag it/them to their respective fields. The same can be done for the folder/s where you would like to have the output deposited. Next, fill all other mandatory field(s) in the input/output block. If an input field is invalid or missing, you will get an error icon ![]() beside the field (to see the error message, you will need to hover the mouse over that icon). Next, configure the form for options and parameters available in the second block of fields. If you set a parameter out of the possible range, you will receive a warning icon
beside the field (to see the error message, you will need to hover the mouse over that icon). Next, configure the form for options and parameters available in the second block of fields. If you set a parameter out of the possible range, you will receive a warning icon ![]() . If you need more information about any input or parameter, click on the exclamation mark ! in front of the input field name.
. If you need more information about any input or parameter, click on the exclamation mark ! in front of the input field name.
Filling the interface block for options and parameters is not mandatory. If you run the program without inputting any parameters and/or condition, DeNovoSeq will execute this program using the default conditions for such a program. Once the job input fields and the parameters have been fulfilled and/or configured, click to start button at the end the interface form to run the Job. If the job has been successfully launched, you will get a confirmation message. Otherwise revise again all input and outputs upload and the options if any.





